Flexible Data Input
Overview
Problem
Users need to enter and specify data for the miRNA target search.
Context
The mRNA or miRNA data input is required to search the database for targets. To narrow the results to the most relevant ones, the data input must allow specifying the search as much as possible. In a miRNA target search, the data input needs to be flexible to accept user data and allow Data Filter.
Solution
Allow the user to enter the data needed to search for miRNA targets in a delimited area, highlighted on the page, and with different advanced search tools. The Flexible Data Input can contain a text box to enter a nucleotide sequence or allow the selection of a sequence from a list predefined by the database. The upload of sequence(s) in a FASTA file must also be supported. The advanced search must allow the user to specify parameters to narrow the search, such as species, a family of miRNAs, and validation method (Data Filter). The tool should give clear examples of how the input is formatted.
Rationale
The starting point of a search tool is data input. Data input should therefore occupy a prominent place in the interface. The target search may start with the target mRNA sequence or the miRNA, so a Flexible Data Input must be available to the user. Lack of flexibility and nomenclature problems are common in miRNA databases (Mullany et al., 2015), as is the difficulty of entering more than one input sequence at a time (Jung et al., 2015). Flexible Data Input allows users to quickly enter textual data, upload their FASTA sequences, select targets from a list predefined by the tool, and specify advanced filters to narrow the search results.
Related patterns
Contains
- Data Filter from Bioinformatics IDP.
Improves
- Input Prompt from UI Patterns.
Examples
miRDB
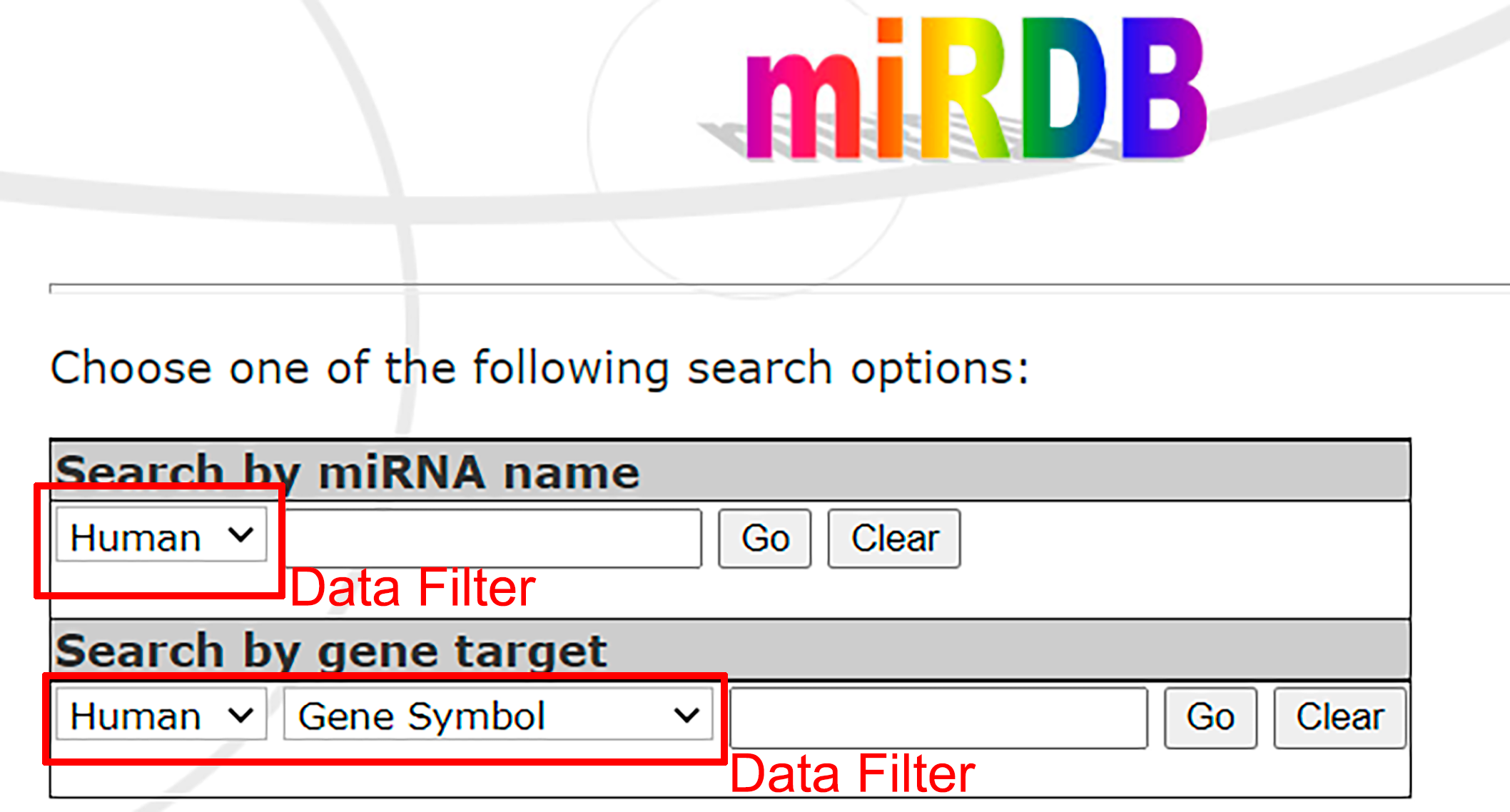
- The interface makes it possible to search starting with miRNA or target gene.
- Data Filter allows you to specify the species and nomenclature.
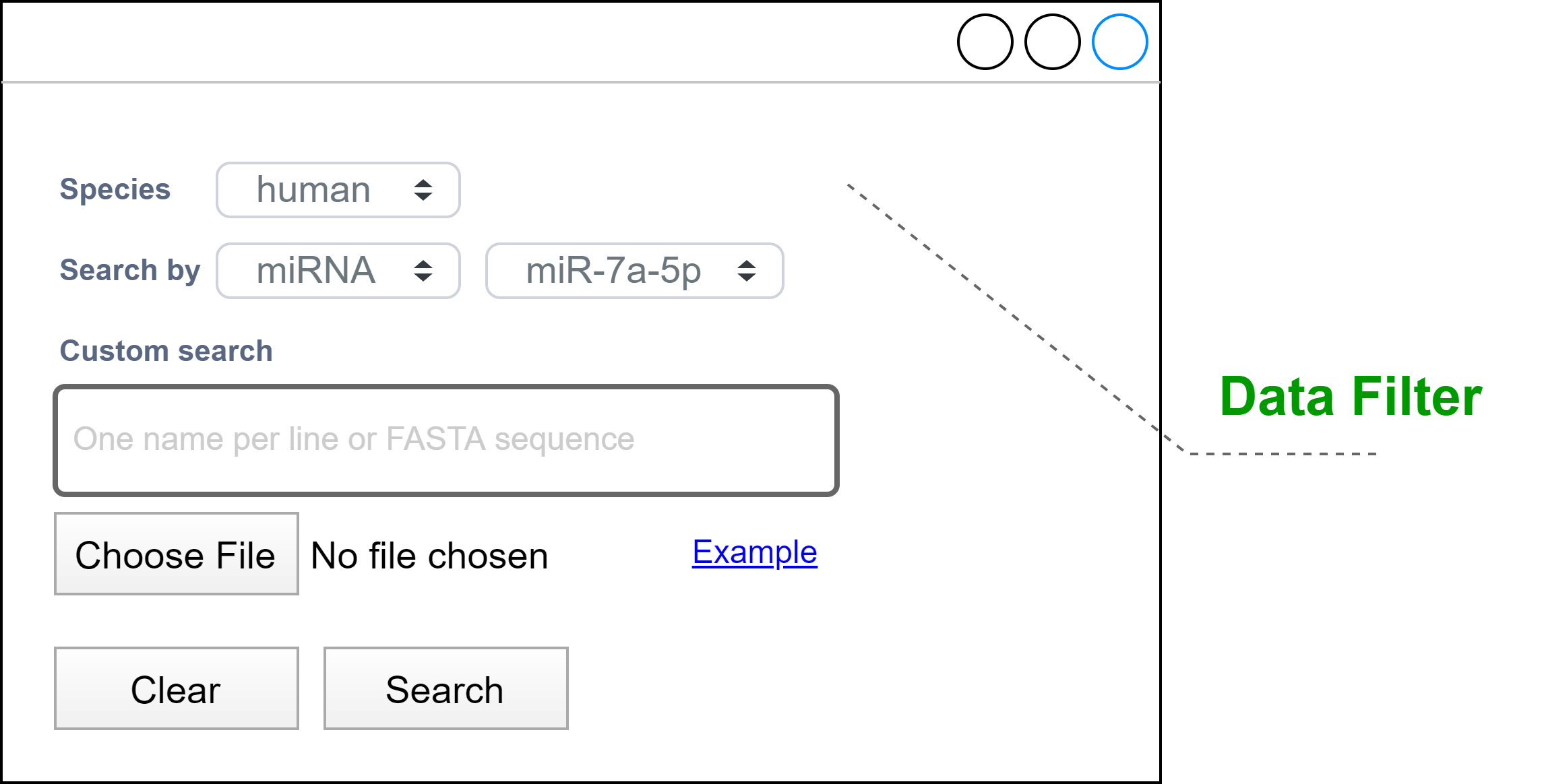
miRWalk
- Search can start with miRNA or gene.
- Allows the user to enter a custom list.
- Clear examples of the data entry pattern.
RNA22
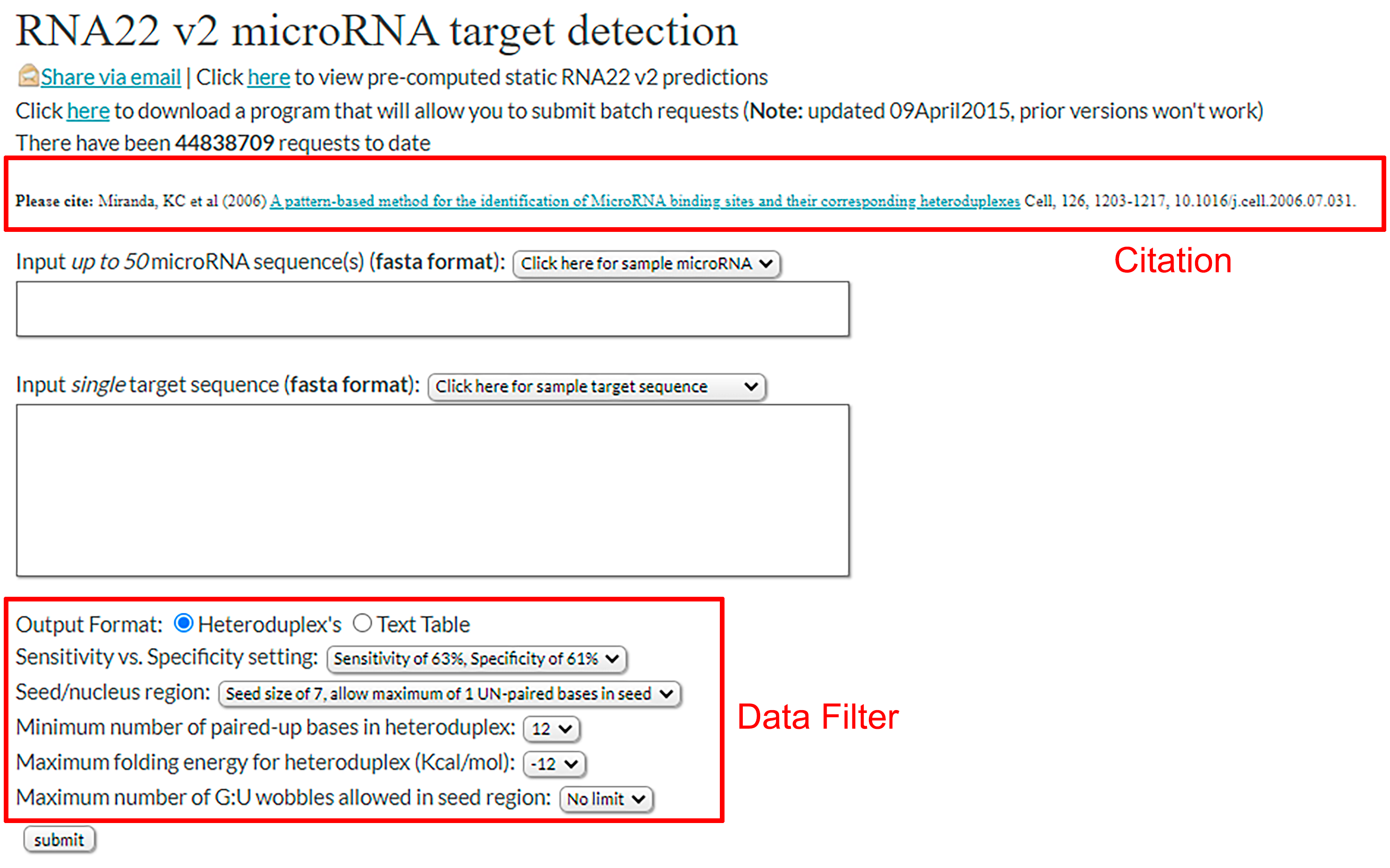
- Possible to insert miRNAs (multiple) or target sequence.
- Examples are also present.
- Data Filter to select the parameters used by the search algorithm.
References
Jung, Daekyoung, et al. “miRTarVis: an interactive visual analysis tool for microRNA-mRNA expression profile data.” BMC proceedings. Vol. 9. No. 6. BioMed Central, 2015.
Mullany, Lila E., Roger K. Wolff, and Martha L. Slattery. “Effectiveness and usability of bioinformatics tools to analyze pathways associated with miRNA expression.” Cancer informatics 14 (2015): CIN-S32716.